Author
Author  Correspondence author
Correspondence author
Plant Gene and Trait, 2010, Vol. 1, No. 1 doi: 10.5376/pgt.2010.01.0001
Received: 28 Jul., 2010 Accepted: 06 Aug., 2010 Published: 06 Sep., 2010
Reuzeau et al., 2005, TraitMill: a Discovery Engine for Identifying Yield-enhancement Genes in Cereals, Molecular Plant Breeding, 3(5): 753-759
Transgenesis is a powerful and effective mode to study plant development. CropDesign has developed the TraitMill platform, a high-throughput technology that enables large-scale transgenesis and plant evaluation. The TraitMill is a highly versatile tool for testing the effect of genes and gene combinations on plant phenotypes. It can be used to successfully evaluate hundreds of independent promoter-genes combinations per year, either under optimal growth conditions or under different abiotic or nutrient stress regimes. The TraitMill platform operates in rice and is specially designed to measure alterations in growth with high sensitivity. To date TraitMill is the only platform that combines these two features and is therefore uniquely placed to identify genes that improve the yield of cereals
Backgroud
For many crops, yield is the primary trait in breeding programs. Yet, till recently, the understanding of yield at the molecular level has been very limited. Probably the most important reason for this lack of the molecular understanding of yield is the difficulties to investigate yield in model organisms such as Arabidopsis, and in the lab or greenhouse environment that is available to most plant molecular biologists. Also, experimental systems that have contributed greatly to the molecular understanding of many biological processes in plants have been of little aid in the study of yield. For example, reverse genetics approaches rely on specific screens for either loss or gain-of-function phenotypes in a particular trait of interest. In the case of yield, loss-of-function would be manifested as growth deficiency, a phenotype that is obviously shared by all essential genes, whether or not directly involved in yield. Gain-of-function would be visible by a yield improvement. Yet, the relative yield increase that may be expected in such a case is probably below the detection/resolution limit of the experimental systems used by plant molecular biologists. Also, plant growth and the contribution of plant growth to harvestable yield are continuous processes, which makes it more difficult to apply time-course based transcriptome and proteome analyses.
Driven by the profound conviction that ‘yield-enhancement’ genes do exist in plants, CropDesign has built a platform that should lead to the identification of such genes. This platform was named TraitMillTM, and it uses rice as a model plant, rather than Arabidopsis. Amongst the world most important staple food are cereals such as rice, wheat and corn, and there is little doubt that rice will function as a much better model for studying yield in cereals than a dicot plant such as Arabidopsis. Furthermore, as for Arabidopsis, the full genome sequence of rice is publicly available, thus greatly facilitating the cloning and classification of genes and gene families. And while, in contrast to Arabidopsis, the genetic transformation of rice still requires a tissue culture step (which adds considerable complexity to the study of a quantitative trait such as yield, see further), transformation efficiency is in general higher than for other cereal crops.
To enable the identification of ‘yield-enhancement’genes, the TraitMillTM platform needs to meet a few key requirements. First, due to the limited understanding of the molecular basis of yield improvement, the platform has to accommodate the testing and validation of many candidate 'yield-enhancement' genes. At the same time, a genome-wide approach in which all plant genes are tested for yield enhancement is unfeasible, due to the complexity of a yield assay and, accordingly, a lack of resources to manipulate thousands of genes in such assay. This means that each candidate gene has been specifically selected and that constructs are designed to target the expression of such gene in the desired plant organ. Second, the yield assay has to be performed in a contained greenhouse environment, but should at the same time resemble agronomic field conditions, so that the greenhouse performance of a transgenic line has a strong predictive value for its yield characteristics in the field. Third, the resolution of the yield screen in the greenhouse should permit to observe relatively small differences in growth and yield parameters; this in contrast to the mutant screens executed by most scientists, which often solely focus on the identification of dramatic phenotypes.
This article provides a description of the TraitMill platform, focussing on the experimental set-up, the parameters that are measured, and the resolution of the screen that is achieved. The relevance of working directly in rice is illustrated by means of the DWARF4 gene, which, when overexpressed, gives opposite effects in Arabidopsis and rice. A follow-up paper will describe the identification of a yield-enhancement gene construct using TraitMill, and will discuss how the improved yield performance has been successfully validated in field-grown rice.
1 Selection and cloning of gene constructs for expression in transgenic rice
Initially, gene selection was focused on cell cycle control. The cell cycle machinery is an ancient and fundamental mechanism for cell division, of which the basics are shared amongst all eukaryotes. In plants, it not only plays a role in determining leaf size and shape by setting both the rate and plane of cell divisions, but it also controls endoreduplication, a DNA amplification process seen in plant storage organs, such as cereal endosperm and tomato pericarp (Beemster et al., 2005; Coelho et al., 2005; Gonzalez et al., 2004). This initial field of interest has meanwhile been broadened to include other regulatory pathways that can lead to enhanced yield and yield stability, such as hormone homeostasis, photosynthesis, and alterations in sink/source strengths (van Camp, 2005).
For cloning, the GatewayTM Cloning Technology is used, which is a system based on DNA recombination (Karimi et al., 2002). With this highly efficient and versatile system, the DNA segments (genes or cDNAs) can easily be transferred between multiple Destination Vectors, harbouring different promoters for driving transgene expression. These promoters were identified through a proprietary promoter discovery program and include promoters with preferential expression in either roots, shoots, meristems, embryo, aleurone, and endosperm.
2 Transgenic plant production
CropDesign uses a proprietary procedure to transform rice plants based on DNA transfer mediated by Agrobacterium tumefaciens. The protocol is a modified version of a transformation method developed by the National Institute of Agrobiological Sciences (Tanaka et al., 2002, Method for superrapid transformation of monocotyledon, Patent application No.: EP 99/0931481). The starting material for this transformation method is mature rice seeds, thus obviating the need for a continuous production line of immature embryos. The procedure is faster and more efficient in comparison with other published protocols (>5 000 independent events per man year). It takes about 7 months/28 weeks for generating transformants, from seed to seed. Improvements in the transformation protocol have reduced the length of the callus phase, resulting in major gains not only with respect to the duration of the process but also in the quality of the transformants, since somaclonal variation is reduced. The transformation protocol has also been optimized towards the production of transgenic plants with a low transgene copy numbe (Figure 1).
|
|
3 Molecular characterization of primary transformants
Primary transformant plants (T0) are characterized molecularly for transgene copy number, immediately after their transfer from the in vitro phase to the greenhouse. Leaf tissue is sampled, DNA extracted and presence and copy number of the T-DNA cassette inserted is determined by quantitative PCR. The plants carrying one copy of the T-DNA, are selected for T1 seed production. Plants with higher copy numbers are discarded. On average, about half of the transformants generated for each construct are maintained. This step allows efficient use of greenhouse space and manpower, since only those plants that are used for T1 evaluation will be carried on till seed production.
Careful molecular characterization of the transgenic plants allows a quality filter to the event selection procedure, reducing the risk of selecting events carrying deletions in the region of the T-DNA where the Gene of Interest is located. Further features of the system relate to data management and reporting tools, which facilitate throughput and increase efficiency and information transfer (Figure 2).
|
|
4 Experimental set-up for yield evaluation
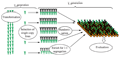
Phenotypic evaluation is performed in the greenhouse on T1 generation plants for five to eight independent events per construct. Several parameters are assessed at different growth stages, from seedlings to mature plants. For each event, half of the T1 plants are null-segregants (azygous) that serve as an internal control to exclude the effects of somaclonal variation (Figure 3).
|
|
Rice plants are grown on a conveyor belt system and most plant handlings are performed by robots. Plants are passed each week through a digital imaging cabinet for measurements on both green and root biomass. Agronomic parameters are extracted from the digital images using various segmentation algorithms. Once plants are mature, seeds are harvested and processed through automated equipment for cleaning, counting and weighing of the seeds. All data are downloaded in a central relational database and automated statistical algorithms are deployed to visualize the performance of transgenic plants versus in-line null segregants. For each construct, an additional analysis is performed by comparing the performance of all the transgenic segregants from different events versus all of the null sister plants of the same evaluation (called the overall effect).
5 Yield parameters
The scored parameters include for example total number of florets per plant, total number of filled seeds per plant, filling rate, thousand kernel weight, seed size, seed dimensions (width and length), number of panicles, number of flowers per panicle, time to flower, maximal aboveground plant biomass, aboveground plant biomass at young stage, harvest index, root biomass, root-shoot index, greenness index. Qualitative parameters such as oil and protein content can also be measured in seeds for specific constructs. The principles for digital analysis of yield-related parameters are exemplified in Figures 4 and 5 for aboveground plant biomass and greenness, respectively.
|
|
|
|
The evaluation of these parameters culminates in the selection of the events in which growth parameters have been most affected, either positively (positive leads) or negatively (negative leads). The seeds of positive leads are then sown and assayed for the presence of the transgene in order to identify homozygous and heterozygous lines. T2 plants of heterozygous lines are subjected to a second evaluation in order to confirm the observed phenotypes. When the trait of interest is observed both in the T1 and in the T2 generation, the transgene effect is considered to be properly inherited and the positive lead is qualified as “validated”. T2 homozygous lines (both with and without transgene) are grown for seed multiplication in view of field trial experiments.
Based on the experience gathered by running the TraitMill over a number of years, CropDesign has optimised its use of resources in view of the desired discrimination power of the yield evaluation. The discrimination power varies for different yield parameters, as determined by the biological variability of the parameter and the accuracy of measuring such parameter. Since seed yield per plant is commercially the most important parameter, the discrimination power of the experimental set-up has been optimised for this trait. From a business perspective, it is anticipated that a yield increase of 10% is required to generate a positive return on investment for a transgenic trait. The TraitMill indeed achieves a ‘least significant difference’(lsd) for a transgene effect on seed yield of 10% at a significance of P<0.05, using a set of approximately hundred plants per construct. With this setup, the lsd at P<0.05 for a transgene effect on above-ground biomass is 7%. For thousand kernel weight, a very stable parameter, the lsd at P<0.05 under the same set-up is 3%. Obviously, the number of plants in an evaluation experiment can be adapted according the desired discrimination power. For example, a higher resolution may be desired for the analysis of inbred lines from a conventional breeding program. The TraitMill platform is indeed also used for evaluation of yield parameters in breeding populations.
6 Lead example
Constitutive overexpression of DWARF4, a gene involved in brassinosteroid signalling, enhances vegetative growth and seed yield in Arabidopsis (Choe et al., 2001). CropDesign has made transgenic rice overexpressing a rice ortholog of DWARF4 under control of the rice GOS2 promoter. Five of these lines were analysed on the TraitMill platform for different yield-related parameters. All five lines showed reduced biomass production and seed yield, as evidenced by different TraitMill parameters. Aboveground biomass, plant height, total number of seeds, total number of filled seeds, total weight of seeds and harvest index were significantly reduced in the transgenic lines versus the non-transgenic controls (significant at P < 0.0001 for the overall data of the five lines). Height was reduced by 7%, aboveground biomass by 13%, height by 7%, total number of seeds by 32% and total weight of seeds by 53% (Figure 6).
|
|
7 Conclusions
CropDesign has built a high throughput platform for rice and has so far tested almost 1 500 transgenic constructs. While slower and more costly as a testing system compared to Arabidopsis, rice has the advantage that is a crop and that it likely serves as a better model for cereal crops than Arabidopsis. Evidence is emerging that the effects of certain genes involved in yield-related developmental processes may be quite different in rice and Arabidopsis (van Camp, 2005). This is further exemplified by the results obtained with DWARF4 overexpression in rice. For this reason, Arabidopsis has been abandoned and all research activities are now focussed on rice.
Over the last years, CropDesign has identified a range of gene constructs that cause interesting phenotypic alterations in transgenic rice. Knowledge of the molecular nature of these genes is now leading to a better understanding of the critical processes that impinge on plant performance. Moreover, the incorporation of root growth into the set of analysed parameters opens new perspectives of gaining insight into the mechanisms that control root development. For obvious reasons, roots have largely been overlooked by breeders, and a better understanding of the contribution and potential of root development for improving yield can now be envisaged.
Interestingly, transgenic lines that in two consecutive generations displayed improved performance for yield parameters such as seed yield or green biomass in the greenhouse, often also showed the same characteristics in the field. This means that the yield data obtained on a per plant basis in the greenhouse have significant predictive value for the performance of the transgenic line in the field, where yield is measured on a per area unit basis. CropDesign's most promising rice leads will be introgressed into more rice varieties for further field testing. Furthermore, the same constructs are being introduced in other crops such as maize and wheat. Analysis of the same constructs in different cereals will generate valuable data on the transferability of yield enhancement strategies from rice to other cereal crops.
References
Beemster G.T.S., de Veylder L., Vercruysse S., West G., Rombaut D., van Hummelen P., Galichet A., Gruissem W., Inzé D., and Vuylsteke M., 2005, Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis, Plant Physiol., 138(2): 734-743 doi:10.1104/pp.104.053884
Choe S., Fujioka S., Noguchi T., Takatsuto S., Yoshida S., and Feldmann K.A., 2001, Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis, Plant J., 26(6): 573-582 doi:10.1046/j.1365-313x.2001.01055.x
Coelho C.M., Dante R.A., Sabelli P.A., Sun Y.J., Dilkes B.P., Gordon-Kamm W.J., and Larkins B.A., 2005, Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in Endoreduplication, Plant Physiol., 138: 2323-2336 doi:10.1104/pp.105.063917
Gonzalez N., Hernould M., Delmas F., Gevaudant F., Duffe P, Causse M., Mouras A., and Chevalier C., 2004, Molecular characterization of a WEE1 gene homologue in tomato (Lycopersicon esculentum Mill.), Plant Mol. Biol., 56(6): 849-861 doi:10.1007/s11103-004-5110-2
Karimi M., Inzé D., and Depicker A., 2002, GATEWAY vectors for Agrobacterium-mediated plant transformation, Trends Plant Sci., 7(5): 193-195 doi:10.1016/S1360-1385(02)02251-3
van Camp W., 2005, Yield enhancement genes: seeds for growth, Curr. Opin. Biotechnol., 16(2): 147-153
doi:10.1016/j.copbio.2005.03.002
. PDF(662KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Christophe Reuzeau
. Jan Pen
. Valerie Frankard
. Joris de Wolf
. Rindert Peerbolte
. Willem Broekaert
. Wim van Camp
Related articles
. TraitMill
. High-throughput technology
. Digital phenotyping
. Functional genomics
. CropDesign
Tools
. Email to a friend
. Post a comment